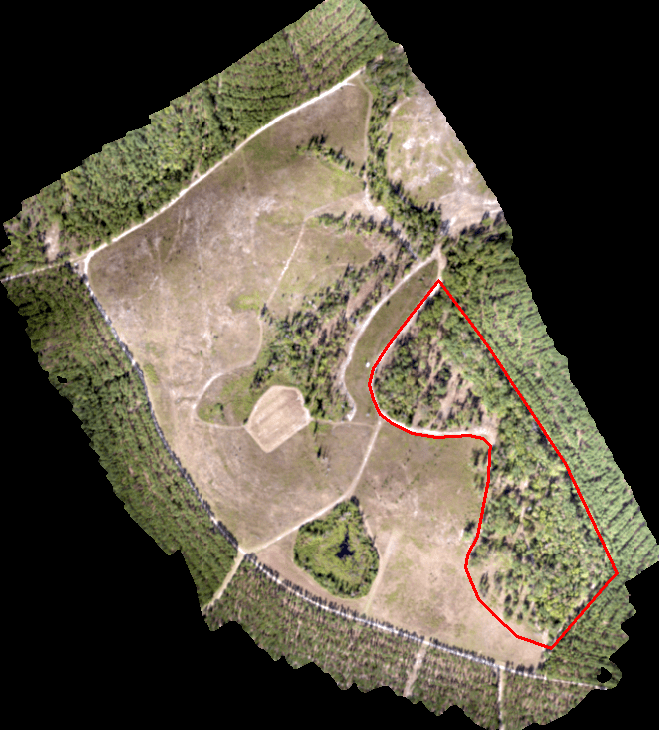
Today’s scenario. We need to perform a similar analysis on a site I flew last summer. We are only interested in the area outlined in red (screenshot below).
INPUT DATA:
- Input LAS: sp2_pcloud.las (DropBox Link)
- Processing extent: AnalysisUnit.shp
R WORKFLOW:
- Clip input data to processing extent
- Classify ground points
- Create bare ground model
- Normalize terrain model
- Create CHM
- Segment image
- Export
LAST WEEK’S CONTENT
#####Code from Lab 9 #####Install/Load libraries source("https://bioconductor.org/biocLite.R") biocLite("EBImage") library(EBImage) library(lidR) library(raster) #####Specify Working Directory #####This should point to the directory in which your *.las resides setwd("C:/temp/Nov30_2017/PhotoscanOutput/") #####Read LAS file #####https://rdrr.io/cran/lidR/man/readLAS.html my_data1<- readLAS("clpSG_PointCloud_th001.las") ##make note of min/max X Y Z: ##make note of Num. of point records summary(my_data1)
NEW CONTENT
#####!!!!!!!!!!!!!!!!!!!!!!!!!!NEW CONTENT!!!!!!!!!! #####Read shapefile pextent<- readOGR( ".","AnalysisUnit") pe<- extent(pextent) xmin<- pe[1] xmax<- pe[2] ymin<- pe[3] ymax<- pe[4] xlist<- c(xmin, xmax, xmax, xmin) ylist<- c(ymax, ymax, ymin, ymin) xylist<- cbind(xlist,ylist) polyextent<- Polygon(xylist) clp_mydat<- lasclip(mydat, polyextent) plot(clp_mydat)
FROM LAB 9 (CLASSIFY GROUND POINTS)
#####Classify ground points #####https://rdrr.io/cran/lidR/man/lasground.html #####ws: sequence of window sizes to be used in filtering ground points ws = seq(0.75,3, 0.75) #####th: sequence of threshold heights above the surface to be considered a ground return #####this step may take some time to run... th = seq(0.1, 1.1, length.out = length(ws)) lasground(my_data1, "pmf", ws, th) plot(my_data1, color = "Classification") #####make a back up of the my_data1 object my_data1_bak<- my_data1
FROM LAB 9 (COMPUTE DTM)
#####compute digital terrain model (DTM), this is the surface minus features sitting on top #####https://rdrr.io/cran/lidR/man/grid_terrain.html dtm1 = grid_terrain(my_data1, res=0.9, method="knnidw", k=10, keep_lowest=TRUE) plot(dtm1) #####process w/ smaller ground resolution my_data1<- my_data1_bak dtm2 = grid_terrain(my_data1, res=0.5, method="knnidw", k=10, keep_lowest=TRUE) plot(dtm2)
FROM LAB 9 (NORMALIZE SURFACE)
#####normalize point cloud: subtract DTM from point cloud; value 0 represents ground-level #####https://rdrr.io/cran/lidR/man/lasnormalize.html lasnormalize(my_data1, dtm2) #####my_data1 now contains the normalized heights ##make note of min/max X Y Z: summary(my_data1) plot(my_data1) my_data1_bak2<- my_data1
FROM LAB 9 (CREATE CHM)
#####create grid canopy #####for each grid, returns the highest point #####https://rdrr.io/cran/lidR/man/grid_canopy.html chm = grid_canopy(my_data1, res=0.25, subcircle = 0.2) plot(chm) my_data1<- my_data1_bak2 chm2 = grid_canopy(my_data1, 0.05, subcircle = 0.025) plot(chm2) #####notice the redefined canopies chm2 = as.raster(chm2) #####smoothing post-process (2x mean) kernel<- matrix(1,3,3) chm2a = raster::focal(chm2, w=kernel, fun=mean) chm2a = raster::focal(chm2, w=kernel, fun=mean) raster::plot(chm2a, col=height.colors(50))
FROM LAB 9 (SEGMENTATION PROCESS)
#####segmentation process #####https://rdrr.io/cran/lidR/man/lastrees.html crowns = lastrees(my_data1, "watershed", chm2a, th=0.25, extra=TRUE) contour = rasterToPolygons(crowns, dissolve=TRUE) tree = lasfilter(my_data1, !is.na(treeID)) plot(chm2, col=height.colors(50)) plot(contour, add=T) #####write out to GIS image raster::writeRaster(crowns, filename="crowns1.tif", format="HFA", overwrite=TRUE)