Sectionals, once again: MzeroA Flight Training – sectionals How to read a VFR sectional chart: https://www.youtube.com/watch?v=NEp1K-TCmD8 Aeronautical Chart User’s guide (Chart symbols): https://www.faa.gov/air_traffic/flight_info/aeronav/digital_products/aero_guide/media/editions/16Jul2020/cug-complete.pdf
FORS7690
If you don’t already have the input data, HERE it is. The clip layer is here: CLIPLAYER. Now that you have a good idea of how the various parameters affect the final output, I have modified the code so that we input the parameters near the beginning of the code […]
#####Code based on https://erinbecker.github.io/r-raster-vector-geospatial/10-vector-csv-to-shapefile-in-r/index.html #####load libraries library("sf") library("raster") #####Set working path, input text file name, output shapefile name mypath<- "~/r-docker-tutorial/lIlk" myfile<- "20201015_1130wetland8.TXT" outfile<- "emi_20201015_1130wetland8.shp" #####Read CSV into a data frame named mydat mydat<- read.csv(file.path(mypath,myfile), na.strings = "NA") nrow(mydat) #####Remove all rows whose longitude value is NA mydat<- subset(mydat, !is.na(mydat$WGS84_LON)) […]
For the second part of your How-To notebook, you will design, execute, assess, revise and repeat a spatial analysis component of your research project. Specifically, I want you to: Identify one spatial analysis component of your research project; those of you working on a non-thesis degree, find a project […]
This R script allows the user to process multiple LAS/LAZ files at once using the catalog functionality of the lidR library. If you look closely, this and the R code from Monday are almost identical. Line 12, here, we create a LAS catalog instead of reading in an individual LAS […]
Using LAZ point cloud linked on our eLC site #####Processing 1 LAZ file. Will generate slope, aspect, and hillshade images; normalize the point cloud; create #####canopy height model and a custom metric #####Save the #####might need to install libraries #####install.packages(lidR) library(lidR) library(rgdal) library(future) plan(multisession, workers = 4L) #####Setting my working […]
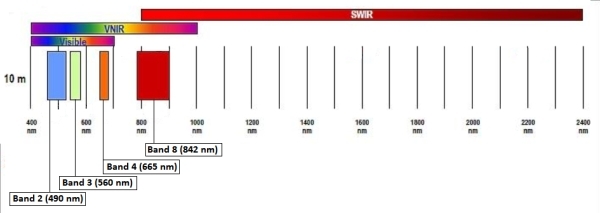
Data: October 13, 2020 Sentinel-2 image (eLC Link) December 18, 2019 Sentinel-2 image (eLC Link) Toolbox (extract the Toolbox.tbx and copy it to your working directory) (eLC Link) Sentinel-2 Satellite Imagery: Instructions for download from USGS Earth Explorer (eLC Link) Data delivered as single-band rasters Sentinel-2 band information 10 meter […]
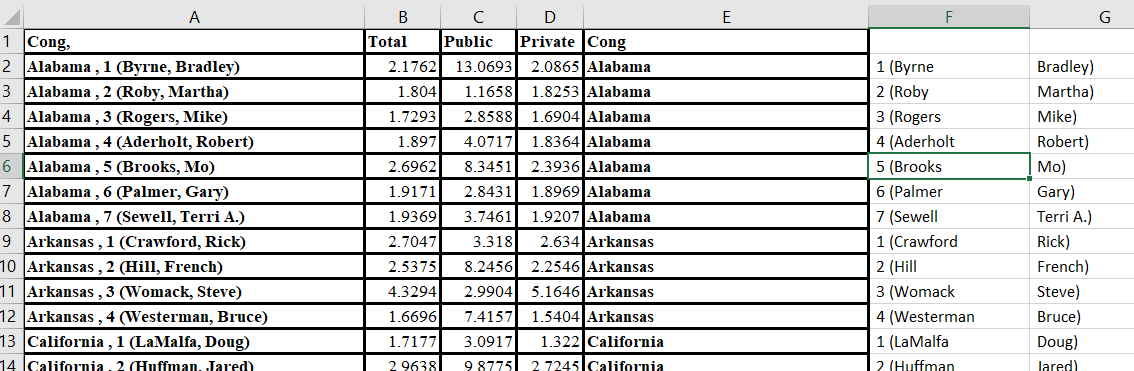
Wednesday, you decided that the GEOID field in the Census’ congressional district shapefile is an appropriate common identifier. I agree, this is a good choice. It is a combination of the state FIPS and the congressional district FIPS. Keep in mind, this should be a four-character code. The data we […]
Forest Inventory and Analysis Program (FIA) Background: The Forest Inventory and Analysis (FIA) Program of the U.S. Forest Service provides the information needed to assess America's forests. As the Nation's continuous forest census, our program projects how forests are likely to appear 10 to 50 years from now. This enables […]
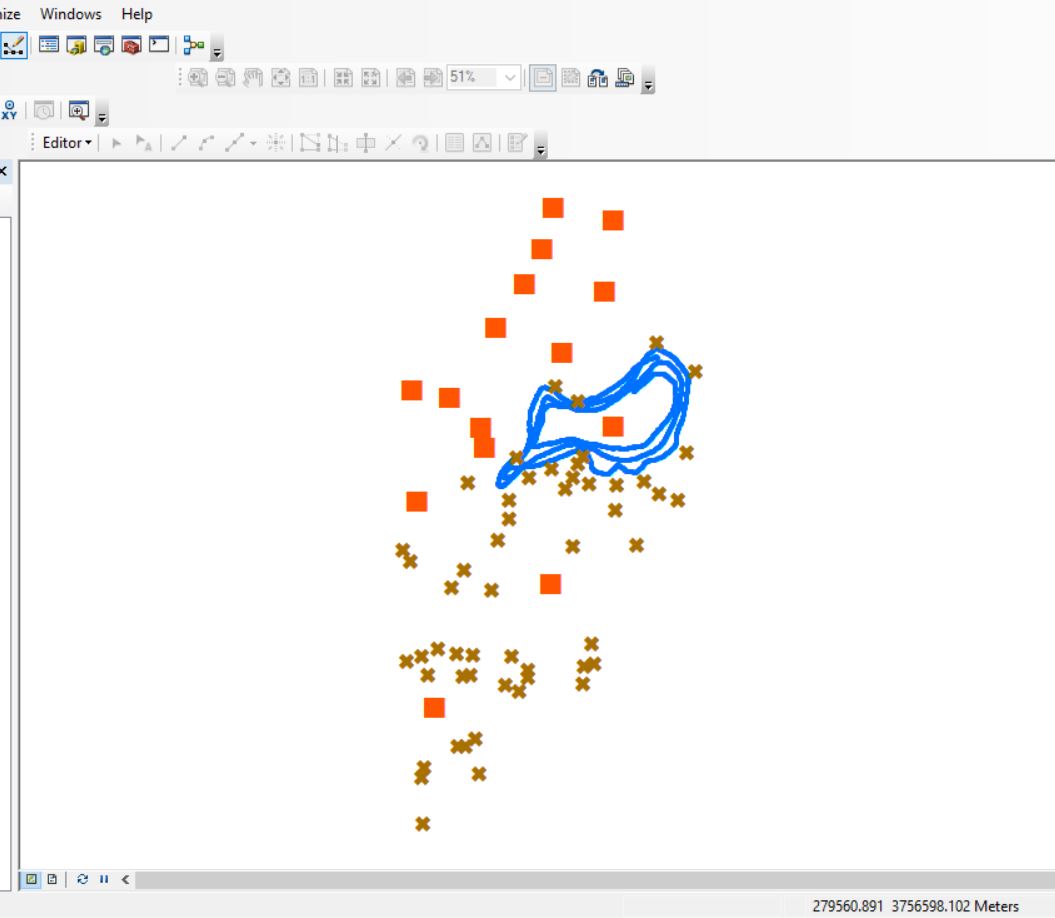
OCONEE FOREST PARK (Lake Herrick): DATA DOWNLOAD LINK Here is the scenario: You have been asked to compile preliminary data for the Oconee Forest Park for an upcoming project. In the download above, you are provided three datasets: Lake_Herrick_Depths.csv: Lake depths captured with a Lowrance fish finder. XY coordinates […]